Biomechanics
|
|
Research Topics:Theoretical modelling; Computational Analysis; Biomechanics; Mutiphysics; Systems (Mechano)Biology; Multiscale Modelling; Patient-specific Modelling; In Silico Medicine Contact: Dr Jérôme Noailly |
RESEARCH | RESSOURCES & INFRASTRUCTURES | PEOPLE | PROJECTS & COLLABORATIONS | JOB OPPORTUNITIES | PUBLICATIONS | COMMUNICATION | ACTIVITIES
RESEARCH
Biomechanics and Mechanobiology (BMMB) is one of the Research Areas of BCN MedTech, led by Jérôme Noailly, co-director of the SIMBIOSys group. It settled at UPF in 2015. Research at BMMB focuses on the load-bearing organs and tissues of the human body in health and disease, and it targets (i) the interactions between tissue multiphysics and biological processes, (ii) the multiscale regulation of organ functional biomechanics, (iii) the identification of mechanistic risk factors associated with different diseases and disorders, based on synthetic data and on the virtual augmentation of real world data.
Numerical methods that combine different modelling and simulation techniques are used to describe both the tissues at the organ level, and the tissue-cell interactions at the tissue and cellular levels. Models are usually developed to admit real world biological and/or clinical data as inputs, in addition to mechanical data from motion analyses where relevant. Theoretical and numerical concepts are tested against in vivo and in vitro data, allowing mechanistic interpretations of both experimental and clinical evidences, in addition to model validations.
On the one hand, emphasis is given in the study of the multiscale transfer of mechanical effects from the system level to the cell level in different scenarios, e.g. simulated treatments, organ/tissue condition or cell cultures; relative to a chosen reference state. On the other hand, advanced tissue models are used to link observable phenotypes to possible mechanisms of spatiotemporal tissue regulation that will depend on the prediction of different cell microenvironments. Both top-down and bottom-up approaches are adopted, to eventually apprehend the regulation of highly multifactorial diseases and disorders.
SPINE & INTERVERTEBRAL DISC
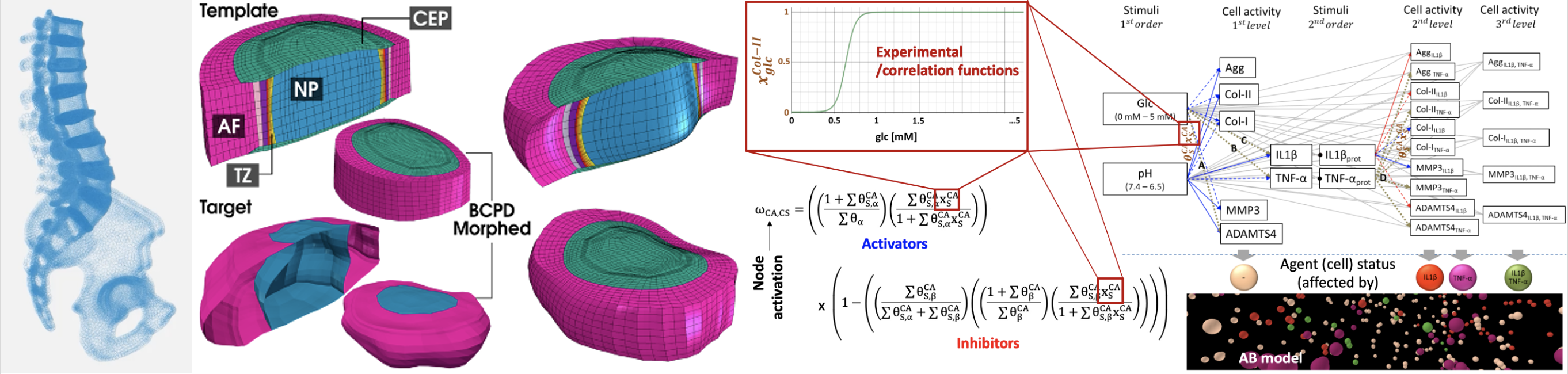
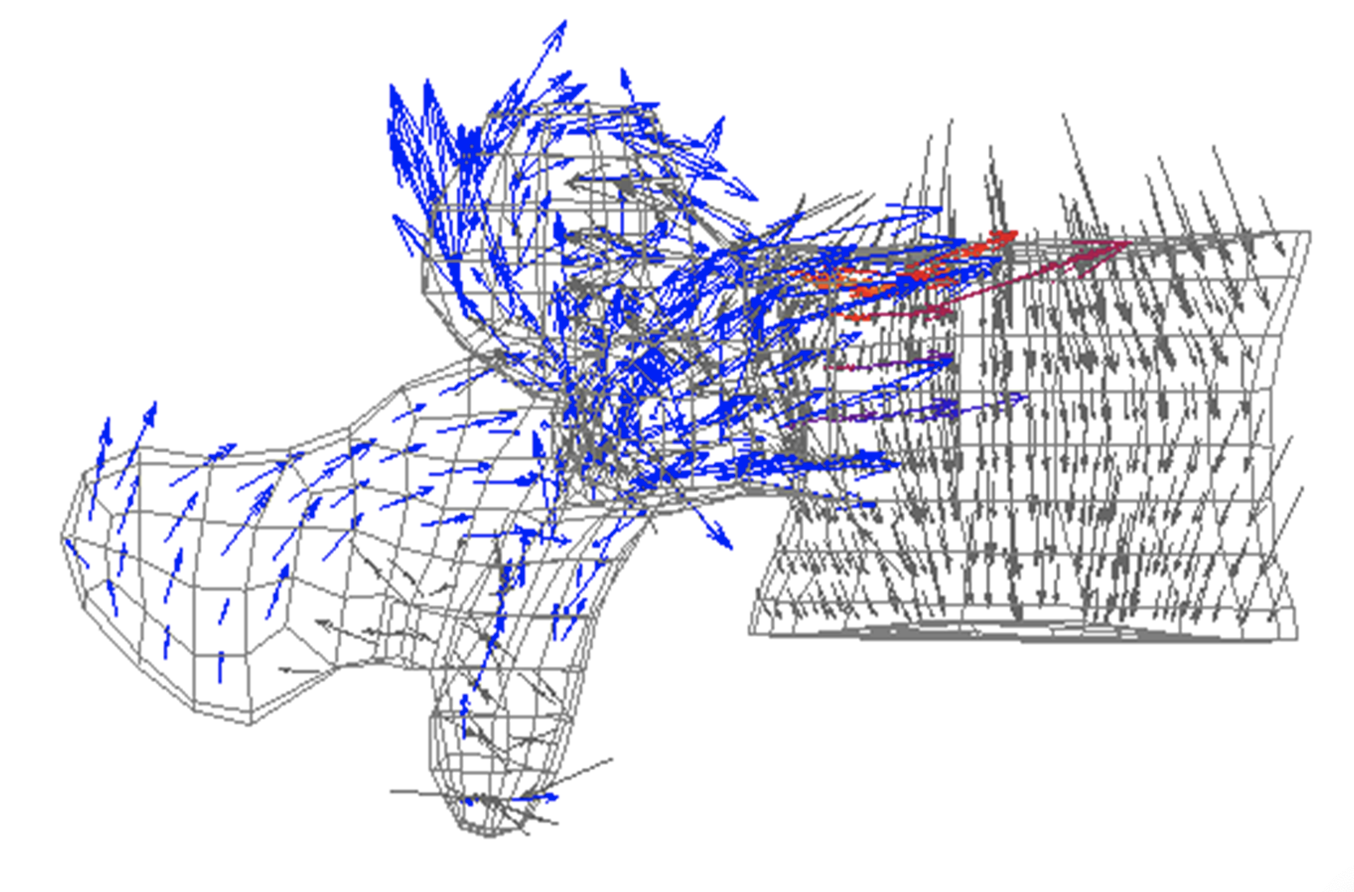
Finite element modelling of the spine and intervertebral disc, for:
- Spine surgery planning and prognosis
- Intervertebral disc pathophysiology exploration
Coupling with:
- image- and point-processing for patient-specific modelling (statistical shape modelling, mesh morphing)
- Tissue multiphysics modelling
- Systems biology models (network, agent-based) for dynamic modelling of intervertebral disc biological regulation
- Subject & biological data (population cohorts, clinical & wet lab collabotations)
| REFERENCE ARTICLE 1 | REFERENCE ARTICLE 2 | REFERENCE ARTICLE 3 |
CARTILAGE & OSTEOARTHRITIS
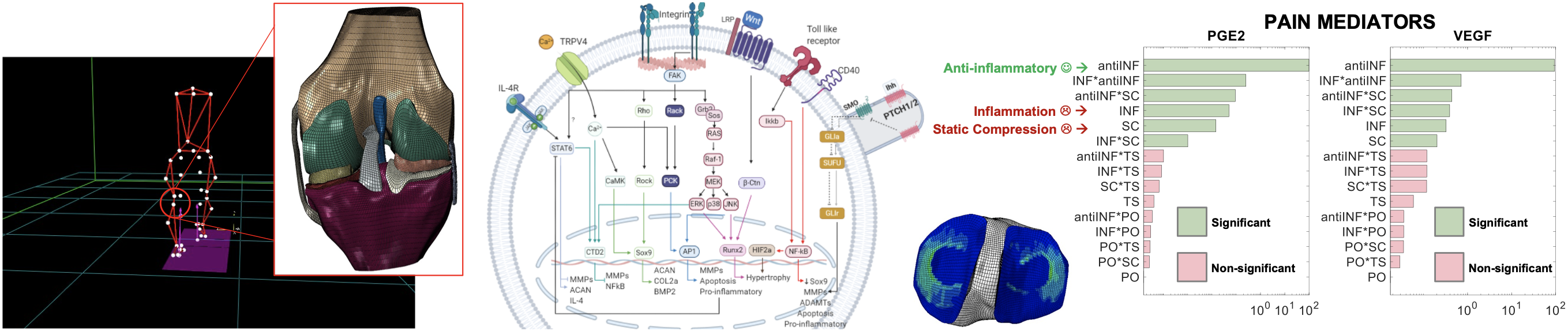
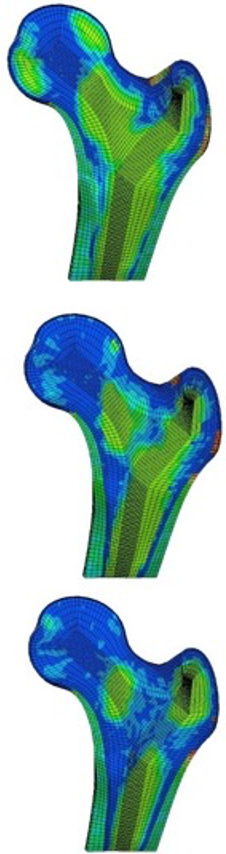
Gait analysis and knee finite element modelling to:
- Capture patient-specific knee joint loads
- Calculate different stress and composition changes in the knee cartilage
Coupled with:
- Osteoarthritis patient clinical and biological data (clinical & wet lab collaborations)
- Tissue multiphysics modelling
- Network models for chondrocyte biochemical regulation coupled to mechanotransduction
| REFERENCE ARTICLE 1 | REFERENCE ARTICLE 2 | REFERENCE ARTICLE 3 |
BONE & OSTEOPOROSIS
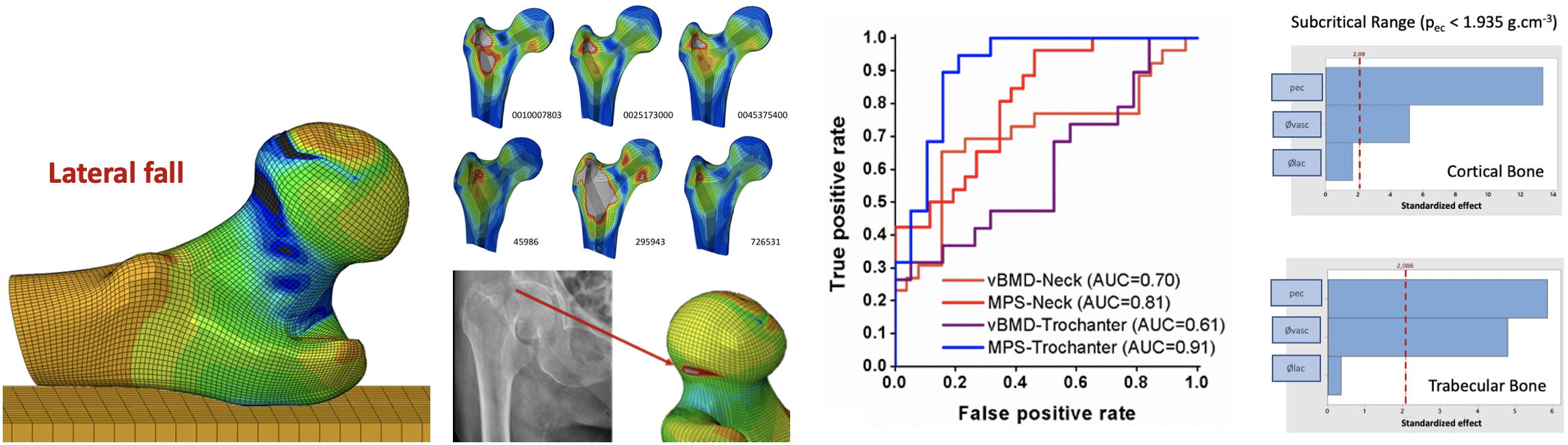
Patient-specific (mesh morphing) and structured finite element modelling to:
- Explore mechanical fracture predictors in osteoporosis patients
- Quantify the mechanical effect of drug treatments in osteoporosis
Coupled with:
- The exploitation of DXA image through the 3D Shaper® technology
- The exploration of the respective contributions of the trabecular and cortical bone tissues
- Different bone constitutive modelling (elastic; elasto-plastic; multi-scale) approaches
MOTION ANALYSIS & BODY FUNCTION
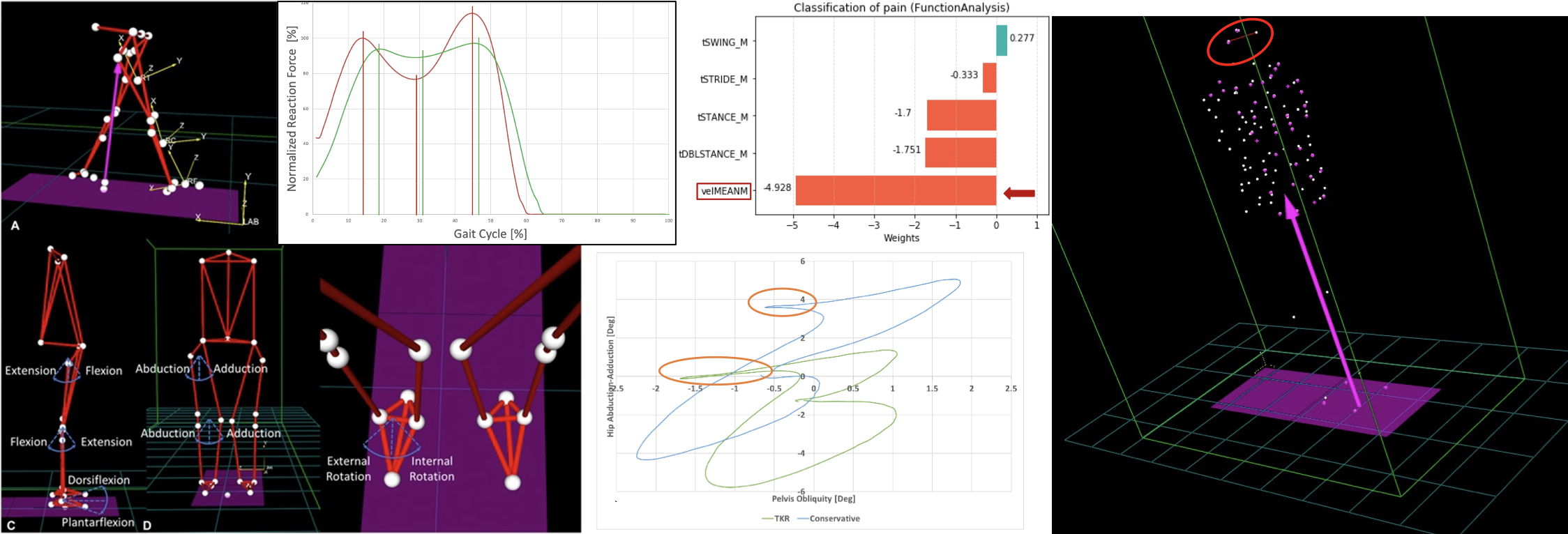
Subject motion & posture capture and analysis to:
- Assess clinical decisions and phenotypes
- Identify descriptors of body stability, e.g., associated with joint coordination, muscle tension or breathing
Coupled with:
- Patient clinical data (clinical collaboration)
- Multivariate statiscal analyses & Machine Learning
- Alternative motion sensor technologies
| REFERENCE ARTICLE 1 | REFERENCE ARTICLE 2 | REFERENCE ARTICLE 3 |
ATHEROSCLEROSIS
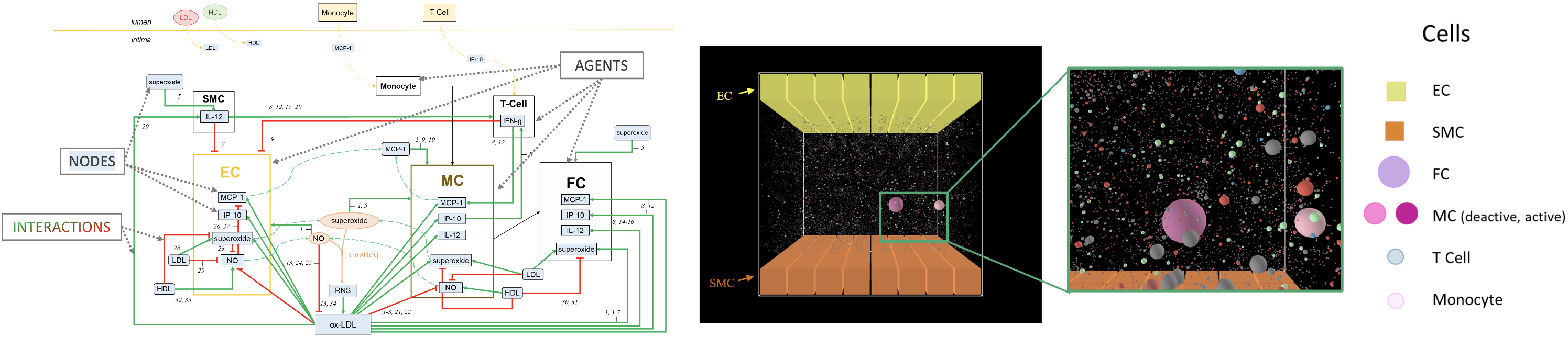
Systems biology, i.e., structured multi-agent-based, network modelling and well-mixed models to:
- Explore the risk factors that shall control the dynamics of atherosclerosis in the sub-clinical phase
- Identify biomarker candidates that inform about plaque development, in terms of both dynamics and type
Coupled with:
- Haemodynamic data
- Molecular biology experiments (wet lab collaborations)
- Patient data (clinical collaborations)
RESPIRATION & CHRONIC OBSTRUCTIVE PULMONARY DISEASE
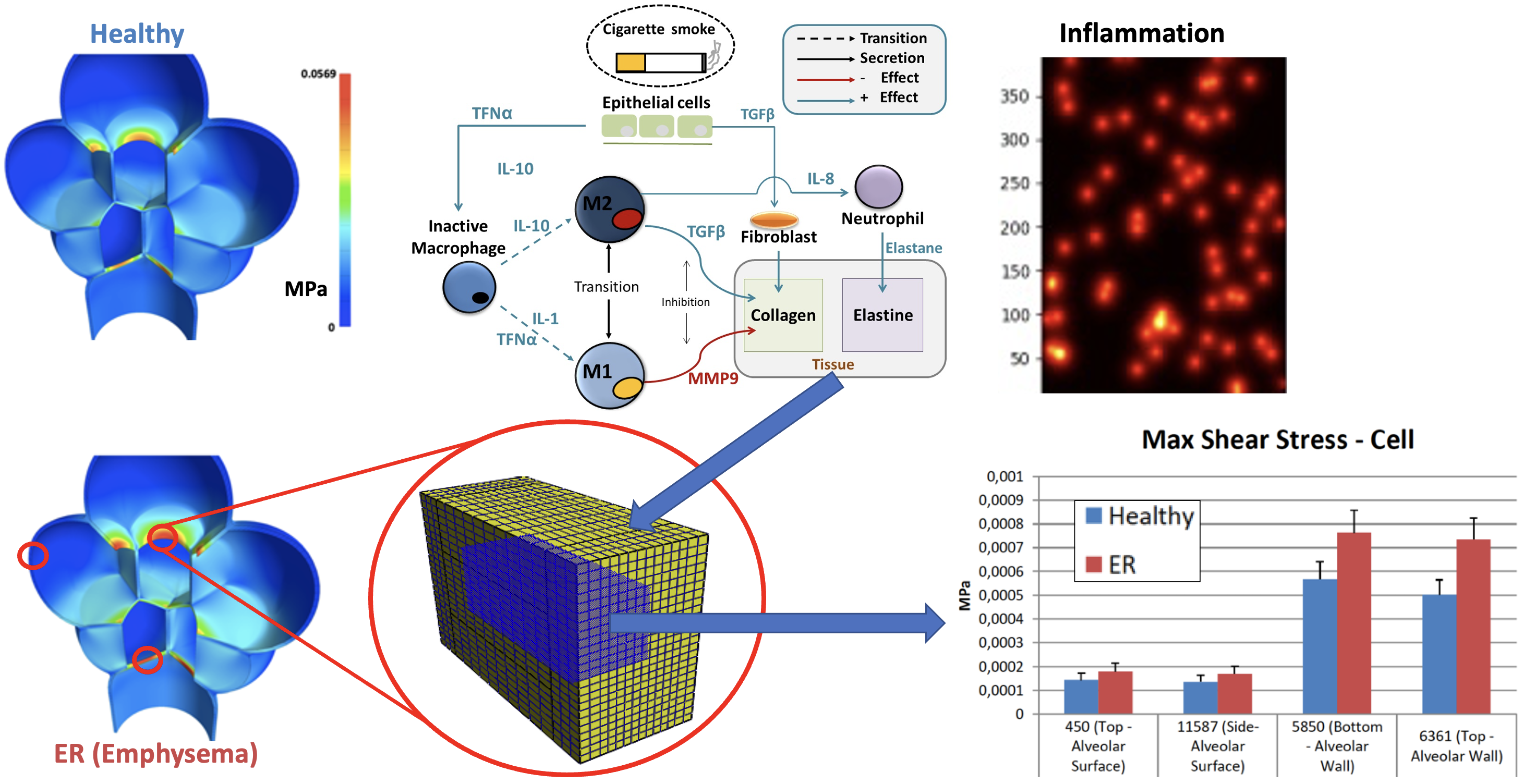
Well-mixed biological and multiscale finite element modelling to:
- Explore the interplay between lung alveolar inflammation and lung parenchyma deformations
- Identify the most relevant mechanobiological interactions that contribute to the destruction of alveaolar walls in emphysema
- Target biological risk factors in lung disease, and early biomarkers for lung emphysema
Coupled with:
- Particle (cigarette smoking, air pullution) inhalation
- Immune system modelling
RESSOURCES & INFRASTRUCTURES
The Biomechanics and Mechanobiology (BMMB) area benefits from different infrastructures and computational ressources, that are both exploited and maintained by teams of professional experts.
The MoCA LAB: MOTION CAPTURE & ANALYSIS
Contact: [email protected]
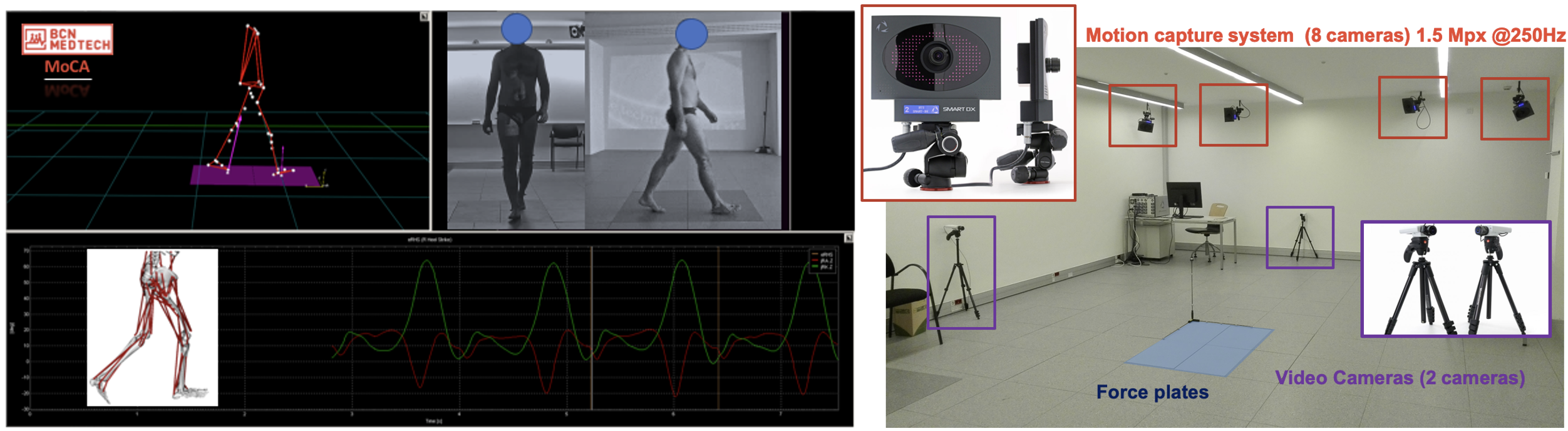
Integrated BTS System with:
- 8 infrared cameras BTS Smart-DX 700: 1.5 MPx resolution; 250 fps; Precision of marker motion of 0.1 mm in a 4x4x3 m volume
- 2 force plates BTS P-6000: 12 sensors /plate; sensitive area 60x40 cm; automatic force calculation; 16 bit resolution; 8000 N capacity in each direction; error < 1% of full scale signal output
- 2 video cameras BTS VIXTA 50 Hz
- 8 Surface EMG BTS FREEMG 1000: 41.5x24.8x14 mm probe dimensions; 13 g probe weight; 2 USB receivers for Real time Wireless IEEE802.15.4 data transmission; 16 bit resolution; 1 kHz acquisition frequency; up to 6 h continous acquisition; data transmission over up to 20 m; solid state storage for up to 1 h stand-alone storage of acquired data
- Smart DX Data Station 32 analogue channels
- Software: BTS SMART Clinic; BTS SMART Analyzer; BTS Digivec
- Stardard protocols: Davis; Helen Heyes; Oxford Foot Model; Bisegmental 3D Foot Helen Heyes; Digivec (Ground Reaction Force Analysis)
FINITE ELEMENT ANALYSIS
- Simulia ABAQUS Floating Licences: 2 ABAQUS CAE; 75 Analysis Credits
- COMSOL Floating Licences: COMSOL Multiphysics; Structural Mechanics Module; Nonlinear Structural Material Module; Porous Media Flow Module; Chemical Reaction Engineering Module
HIGH-PERFORMANCE COMPUTING
As all the research group of the UPF Department of Information and Communication Technologies (DTIC), the BMMB has access to the High Performance Computing Cluster SNOW, the performance of which is in constnt evolution according to the needs of the DTIC Researchers (see current configuration). Currently, BMMB simulations run in Docker and Kubernetes container solutions, for maximum flexibility and efficiency.
PEOPLE

| HEAD |
| POSDOCTORAL RESEARCHERS |
Carlos Ruiz Wills | Laura Baumgartner | Zerihun Workineh | Elham Alizade | Jakka Prasad | Morteza Rasouligandomani | Corine Post
| PhD CANDIDATES |
Alba Fisher | Mayra Loayza | Andreu Pascuet | Jorge Mateos Arriola | Estefano Muñoz | Sofia Tseranidou | Mané Sargsyan | Aylin Kadkhodamanesh
| PROJECT MANAGER |
Aneta Antonova Toncheva | Pablo Orons
| FORMER MEMBERS |
Maria Segara-Queralt | Simone Tassani | Themis Toumanidou | Andy Olivares
PROJECTS & COLLABORATIONS
CURRENT PROJECTS
|
FUNDING:
Ministerio de Ciencia e Innovación y la Agencia Estatal de Investigaciónllllllllll |
BONE STRENGTH (CPP2021-008574)
BONE STRENGTH focusses on the prediction of fragile bone fractures in osteoporosis, out of routine clinical explorations. It is a collaboration between the technology-based company 3D Shaper Medical SL, and BCN MedTech. Osteoporosis is associated with the occurrence of fragile fractures in the elderly population, resulting in a huge burden on the society and the healthcare system. The current clinical gold standard for fracture risk assessment is based on the aBMD measured using DXA scan. However, it fails to identify up to 50% of the patients eligible for bone-specific pharmacological treatments, resulting in fractures that could be prevented. Bone strength, estimated using QCT-based FEA, has been shown to improve osteoporosis diagnosis and management. But, increased exposure of patients to X-ray radiations is a limitation. 3D-Shaper Medical SL has developed a software (3D-Shaper®) that provides clinicians with QCT equivalent parameters without performing a CT scan. BONE STRENTH aims to develop a FEA module for 3D-Shaper® and enable QCT-equivalent bone strength estimations from DXA scans. 3D-Shaper® FEA will be used to evaluate the effect of pharmacological treatments, enabling clinical trials with a reduced economic cost and without performing CT scans. PARTNERS: |
| llllllllllllllllllllllllll |
Training network to advance integrated computational simulations in translational medicine, applied to intervertebral disc degeneration. |
Past projects
|
llllllll llllll FUNDING:
Ministerio de Ciencia e Innovación y la Agencia Estatal de Investigaciónllllllllll |
ANDAMIO (RTC2019-007413-1) will predict, from 2D DXA images, the risk of osteoporotic fracture in 3D in the lumbar spine. The predictive algorithm will rely on biomechanical simulations that integrate the bone morphology and quality, external and internal loads, and the influence of soft tissues. The software will integrate the 3D-SHAPER technology, and use 3D-SHAPER outputs to:
The ultimate clinical goal for ANDAMIO is to predict at least 80% of osteoporotic fractures. In the case of successful clinical validation, the technology will be integrated into a commercial software version that will be personalized to be compatible with machines of the main leaders of the market. |
| llllllllllllllllllllllllll |
It is a multi-disciplinary project that aims to describ the probability to develop musculoskeletal conditions related to physical and psychological stress. |
|
llllllllllllllllllllllllll |
Clinical and virtual examination of patients for holistic and objective description of the osteoarthritis progression mechanisms. |
JOB OPPORTUNITIES

OPEN POSITIONS
**************************************************
-
Senior postdoctoral researcher in computer methods for ERC Consolidator Grant O-Health - DETAIL & EARLY EXPRESSION OF INTEREST
**************************************************
Should you be interested in joining us, feel free to send us a declaration of interest, along with your CV (See group contact).
PUBLICATIONS