Dynamical Systems Biology
Jordi Garcia Ojalvo
Group website
Research Outline
The Dynamical Systems Biology laboratory studies the dynamics of living systems, from unicellular organisms to human beings. We use dynamical phenomena to identify the molecular mechanisms of biological processes such as cellular decision-making and spatial self-organization. Using a combination of theoretical modelling and experimental tools including time-lapse fluorescence microscopy and microfluidics, we investigate dynamical phenomena such as pulses and oscillations, and study how multiple instances of these processes coexist inside cells and tissues in a coordinated way. At a larger level of organization, we use mathematical models of neurons and neural tissues to link the structural properties of brain networks with their function.
Research Lines
Cellular computation
We study how cells process external information, by analyzing how the architecture of their regulatory networks allow them to encode the recent history of the cell and predict future changes in their environment.
Stress response in bacteria at the single-cell level
We investigate how bacterial cells self-organize in space and time when subject to different kinds of stresses, including nutrient starvation and antibiotics.
Dynamics of cellular signaling
We analyze the temporal organization of signaling processes at the single-cell level in organisms ranging from yeast to mammalian cells. We also study the regulation of the immune response in the context of autoimmune diseases such as multiple sclerosis.
Brain dynamics
We study the dynamics of the brain at different scales, from the microscopic scale of neuronal networks to the macroscopic scale of the full brain, in the context of a variety of pathologies including Down's syndrome and Alzheimer's disease.
Team during 2019-20
PhD students: Leticia Galera Laporta, Lara Escuaín de Poole, Carlos Toscano Ochoa, Keith Kennedy, Gabriel Torregrosa Cortés, Maria Kokkaleniou, Pablo Casaní Galdón, Alda Sabalic
Postdocs: Gang Zhou, Leila Cababie
Technicians: Miquel Sas Gil, Cristina Fernández Avilés
Selected publications
Galera-Laporta, L., & Garcia-Ojalvo, J. (2020). Antithetic population response to antibiotics in a polybacterial community. Science Advances, 6(10), eaaz5108. https://doi.org/10.1126/sciadv.aaz5108
Lee, D. D., Galera-Laporta, L., Bialecka-Fornal, M., Moon, E. C., Shen, Z., Briggs, S. P., Garcia-Ojalvo, J., & Süel, G. M. (2019). Magnesium Flux Modulates Ribosomes to Increase Bacterial Survival. Cell, 177(2), 352-360.e13. https://doi.org/10.1016/j.cell.2019.01.042
Matsuda, M., Hayashi, H., Garcia-Ojalvo, J., Yoshioka-Kobayashi, K., Kageyama, R., Yamanaka, Y., Ikeya, M., Toguchida, J., Alev, C., & Ebisuya, M. (2020). Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science, 369(6510), 1450–1455. https://doi.org/10.1126/science.aba7668
Saiz, N., Mora-Bitria, L., Rahman, S., George, H., Herder, J., Garcia-Ojalvo, J., & Hadjantonakis, A.-K. (2020). Growth factor-mediated coupling between lineage size and cell fate choice underlies robustness of mammalian development. ELife, 9, e56079. https://doi.org/10.7554/eLife.56079
Yang, C.-Y., Bialecka-Fornal, M., Weatherwax, C., Larkin, J. W., Prindle, A., Liu, J., Garcia-Ojalvo, J., & Süel, G. M. (2020). Encoding Membrane-Potential-Based Memory within a Microbial Community. Cell Systems, 10(5), 417-423.e3. https://doi.org/10.1016/j.cels.2020.04.002
Other relevant information
-
Invited talks in the following conferences:
-
Int. Conference on Quantitative Biology, Yantai, China, June 23, 2019
-
Statphys 27, Buenos Aires, July 11, 2019
-
Annual meeting of the German Physical Society, online, March 18, 2020
-
-
Invitations for research stays in other institutions:
-
University of California San Diego, January 13-19, 2020
-
California Institute of Technology, March 6-14, 2020
-
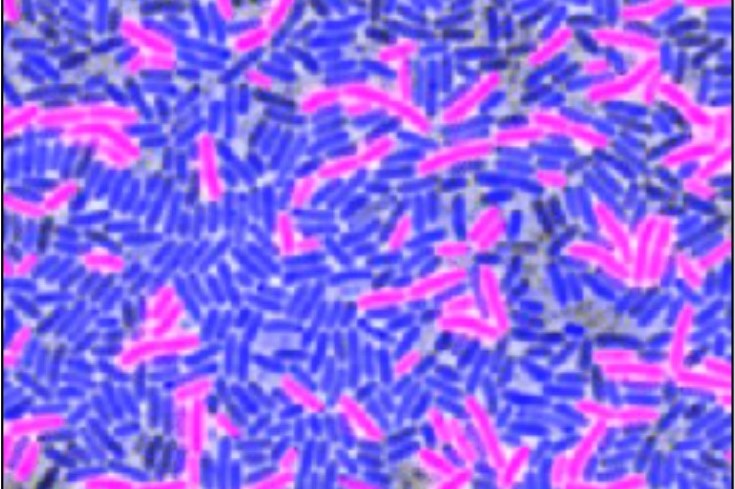
Co-culture of B. subtilis (magenta) and E. coli (blue) bacteria under low antibiotic stress
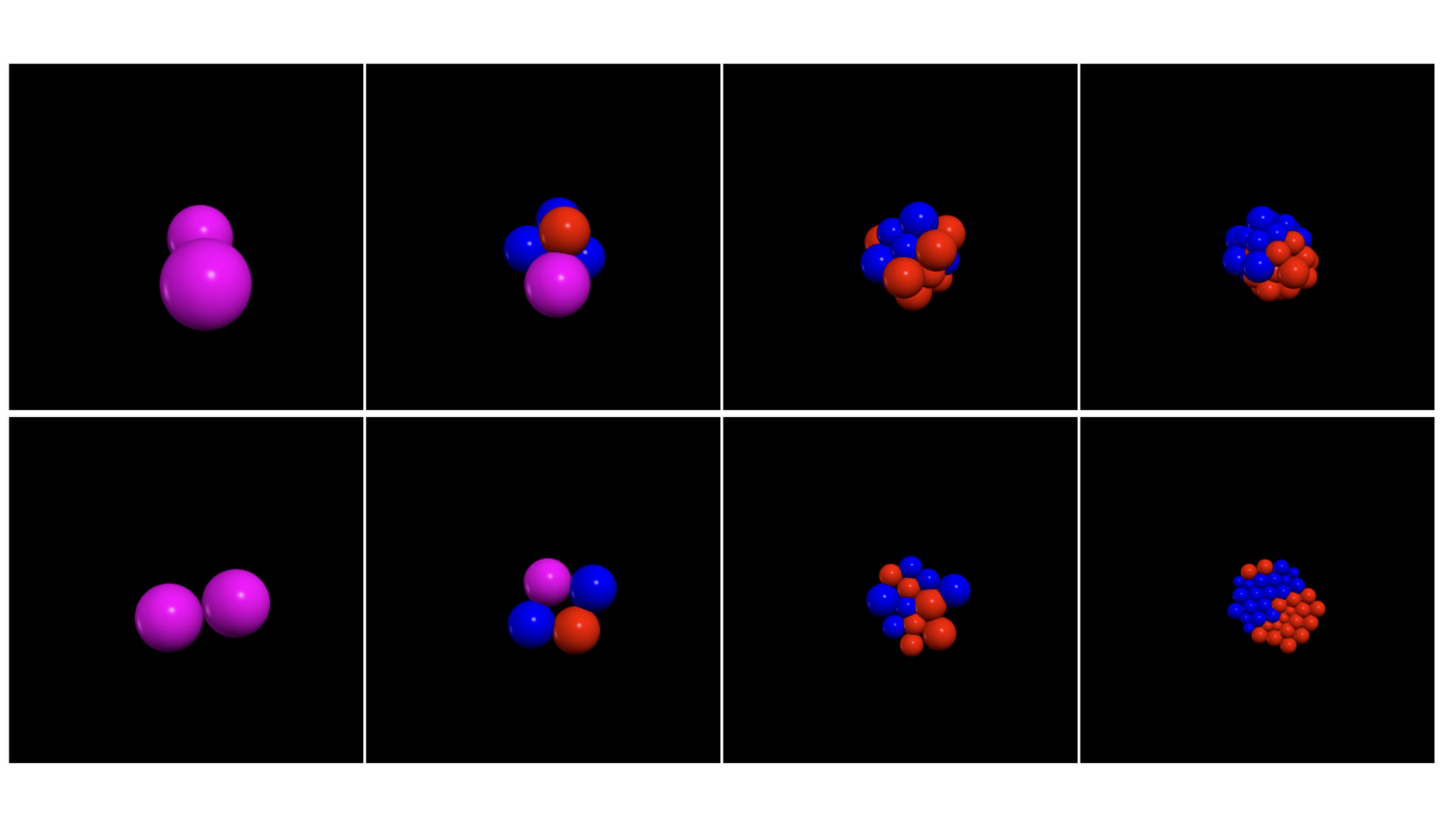
Agent based simulation of the cell-fate decision between epiblast (red cells) and primitive endoderm (blue cells) in the early mouse embryo (top row), and in a two-dimensional disk of pluripotent cells (bottom row).
