Genomics
Ferran Casals
Research Outline
One of the missions of the Genomics Core Facility is the development of new technologies and methodologies for genomic analyses. The Genomics Core Facility at the UPF performs collaborative research projects with labs at the Experimental and Health Sciences Department of the UPF, to develop and implement novel methodological approaches. The laboratory also develops its own research projects.
UPF Collaborative Projects 2020-2021
UPF Collaborative Projects 2018-2019
Services
The Genomics Core Facility at the UPF provides a wide variety of methods for DNA and RNA analyses. Available equipment includes liquid handling robots to automate pipetting tasks, capillary sequencers for Sanger sequencing and fragment analysis, DNA quantification and quality control with Picogreen and Bioanalyzer, real-time PCR and OpenArray system for absolute and relative quantification of nucleic acids (genotyping and gene expression), and four next-generation sequencing platforms: MiSeq (Illumina), ideal for targeted and small genome sequencing; MiSeqFGx (Illumina), for population and forensic genetics analyses; NextSeq (Illumina), a highly flexible platform performing a broad range of applications, from targeted resequencing to RNA profiling and whole-exome or genome sequencing.; and MinION (Oxford Nanopore), a new single-molecule technology ideal for de novo sequencing and the study of structural variation.
The laboratory also offers the service of library preparation for most of the next-generation sequencing applications. The Genomics Core Facility organizes courses and workshops on the new sequencing methodologies and pioneers some of their applications to medical and population genomics research.
Team during 2019-20
- PhD students: Laura Batlle Masó, Manuel Solís-Moruno, Núria Bonet Martín
- Technicians: Roger Anglada Busquets, Núria Bonet Martín, Raquel Rasal Soteras, Marc Tormo Puiggròs
Selected publications 2019-20
• Kuderna L, Solís-Moruno M, Batlle-Masó L, Julià E, Lizano E, Anglada A, Ramírez E, Bote A, Tormo M, Marquès-Bonet T, Fornas O, Casals F. (2020) Flow sorting enrichment and nanopore sequencing of chromosome 1 from a Chinese individual. Frontiers in Genetics, 10, 1315.
• Batlle-Masó L, Mensa-Vilaró A, Solís-Moruno M, Marquès-Bonet T, Aróstegui JI, Casals F.(2020). Genetic diagnosis of autoinflammatory disease patients using clincal exome sequencing. European Journal of Medical Genetics, 63(5), 103920.
• Costa D, Bonet N , Solé A, González de Aledo-Castillo JM, Sabidó E, Casals F, Rovira C, Nadal A, Marin JL, Cobo T, Castelo R.(2020). Genome-wide postnatal changes in immunity following fetal inflammatory response. The FEBS journal, Oct 2.doi: 10.1111/febs.15578.
• Harding T, Milot E, Moreau C, Lefebvre JF, Bournival JS, Vézina H, Laprise C, Lalueza‐Fox C, Anglada R, Loewen B, Casals F, Ribot I, Labuda D. Historical human remains identification through maternal and paternal genetic signatures in a founder population with extensive genealogical record. American Journal of Physical Anthropology, 2020, 171(4), 645-658.
• Barrio PA, García O, Phillips C, Prieto L, Gusmão L, Fernández C, Casals F, Freitas JM, González-Albo MC, Martín P, Mosquera A, Navarro-Vera I, Paredes M, Pérez JA, Pinzón A, Rasal R, Ruiz-Ramírez J, Trindade BR, Alonso A. The first GHEP-ISFG collaborative exercise on forensic applications of massively parallel sequencing. Forensic Sci Int Genet . 2020 Sep 18;49:102391.
Other relevant information 2019-20
Course RNASeq- How low can you go?. Organized by Werfen & New England Biolabs & Genomics Core Facility UPF. At the UPF, 27-29 May 2019.
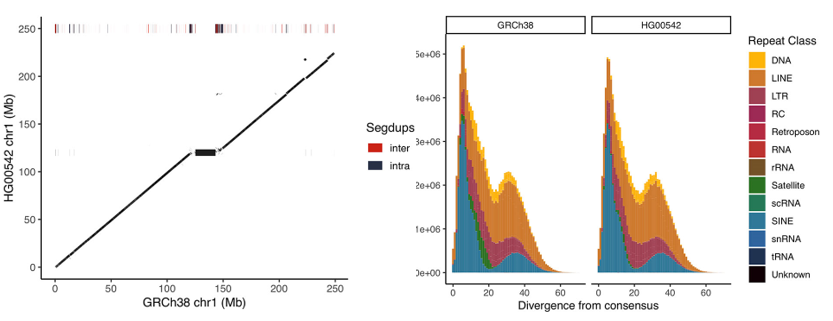
Flow sorting enrichment and nanopore sequencing of chromosome 1 from a Chinese individual. Frontiers in Genetics, 10, 1315.
