Evolutionary Population Genetics
Elena Bosch
Group website
Research Outline
Our research focuses on understanding (i) how adaptive natural selection has shaped the genetic and phenotypic variation in present-day human populations; and (ii) the evolutionary trade-offs explaining some common genetic predispositions to complex disease. For that, we usually analyze genomewide data from different case/control settings or geographically diverse populations and apply state-of-the-art methods for variant association and detection of selection. Moreover, we also aim to elucidate the genetic variants and molecular phenotypes underlying the genetic basis of adaptations presumably related to pathogen interaction and diet by using in silico functional predictions and relevant molecular biology techniques.
Research Lines
Adaptation to zinc deficiency
We are investigating whether human populations living in zinc-deficient soil areas carry signatures of polygenic selection on the whole set of zinc transporter genes. Moreover, we are testing whether particular variants linked to such signatures can be associated with differential zinc content or transport.
Adaptations in the New World
We are analysing whether Peruvian populations from different environments (coast, Amazonian jungle, and Andean highlands) present contrasting local adaptations as well as shared adaptive signals with other Native American populations resulting from their recent common history.
Adaptations related to the pygmy phenotype
We are investigating whether the signature of adaptation found around a non-synonymous variant in the Andamanese is shared by other SE Asian pygmy groups. In addition, we are interrogating the putative adaptive phenotype of this variant on a mouse model to understand whether it could contribute to the Pygmy phenotype.
Team during 2019-20
PhD students: Ana Roca-Umbert, Barbara Sinigaglia, Rocío Caro Consuegra
Technicians: Mònica Vallès
Selected publications
-
Rodríguez JA, Farré X, Muntané G, Marigorta UM, Hughes DA, Spataro N, Bosch E*, Navarro A* (2019). Reply to: Retesting the influences of mutation accumulation and antagonistic pleiotropy on human senescence and disease. Nature Ecology & Evolution 3: 994-995. doi: 10.1038/s41559-019-0926-y. PMID: 31235924.
-
Urnikyte A, Flores-Bello A, Mondal M, Molyte A, Comas D, Calafell F, Bosch E*, Kucinskas V* (2019). Patterns of genetic structure and adaptive positive selection in the Lithuanian population from high-density SNP data. Scientific Reports. 9(1):9163. doi: 10.1038/s41598-019-45746-3. PMID: 31235771.
-
Muntané G, Farré X, Bosch E, Martorell L, Navarro A, Vilella E (2020). The shared genetic architecture of schizophrenia, bipolar disorder, and lifespan. Human Genetics. doi: 10.1007/s00439-020-02213-8. PMID: 32772156.
-
Dobon B, Ter Horst R, Laayouni H, Mondal M, Bianco E, Comas D, Ioana M, Bosch E, Bertranpetit J, Netea M (2020). The shaping of immunological responses through natural selection after the Roma Diaspora. Scientific Reports 10:16134. doi: 10.1038/s41598-020-73182-1. PMID: 32999407.
-
Walsh S, Izquierdo-Serra M, Acosta S, Edo A, Lloret M, Moret R, Bosch E, Oliva B, Bertranpetit J, Fernández-Fernández JM (2020). Adaptive selection drives TRPP3 loss-of-function in an Ethiopian population. Scientific Reports 10:20999. doi: 10.1038/s41598-020-78081-z. PMID: 33268808.
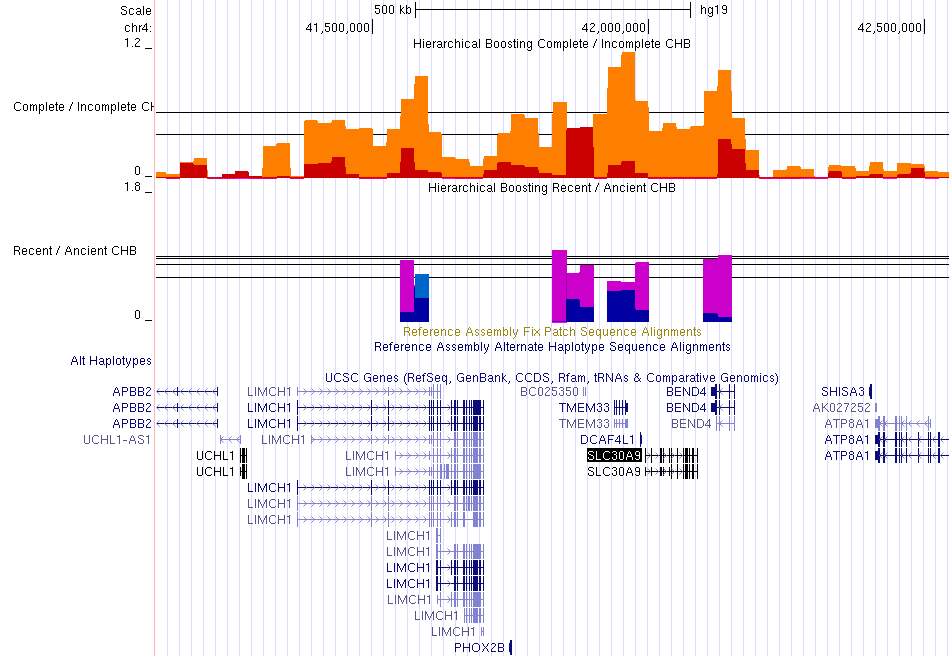
Signatures of positive natural selection around the SLC30A9 gene.
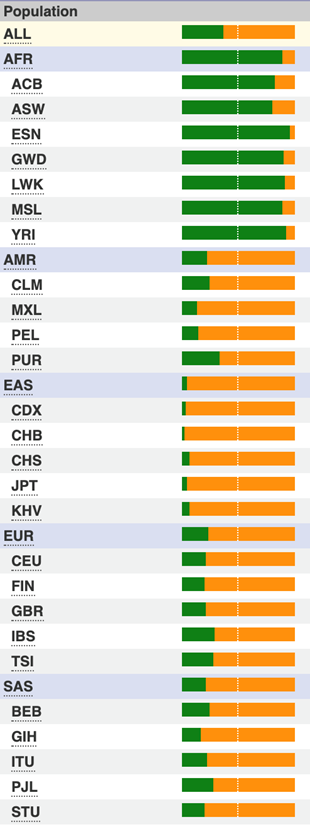
Allele frequencies of the rs1047626 SNP showing high population differentiation between African (top) and non-African populations.
