Image segmentation
Segmentation of medical images is an essential pre-processing step for many image analysis task. Our group mainly focuses on the segmentation of brain images, although our methodologies can be also applied to other types. Two main application areas of our methods are 1) the analysis growth patterns in the developing brain and 2) the understanding of degeneration processes underlying Alzheimer's disease (see applications).
Multiple Atlas Segmentation
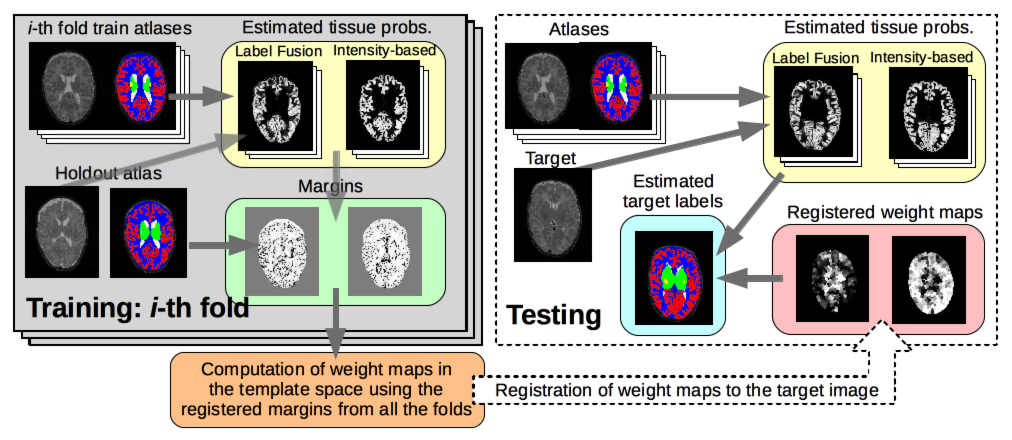
Our research interests are focused on a recent segmentation technique called Multiple-Altas segmentation (MAS). In MAS, we use a set of atlas images along with their corresponding labelmaps to segment a new target image. The atlas labelmaps, denoting the structures to be segmented, are usually obtained by manual or semi-automatic expert annotation. The process of MAS consists of 3 steps: 1) atlas selection, where the most suitable atlases for a given target image are selected, 2) image registration, where the atlas images and labelmaps are warped so as to be in correspondene with the to-be-segmented target image and 3) label fusion, where the decisions from the multiple warped atlas labelmaps are fused into a consensus segmentation on the target image.
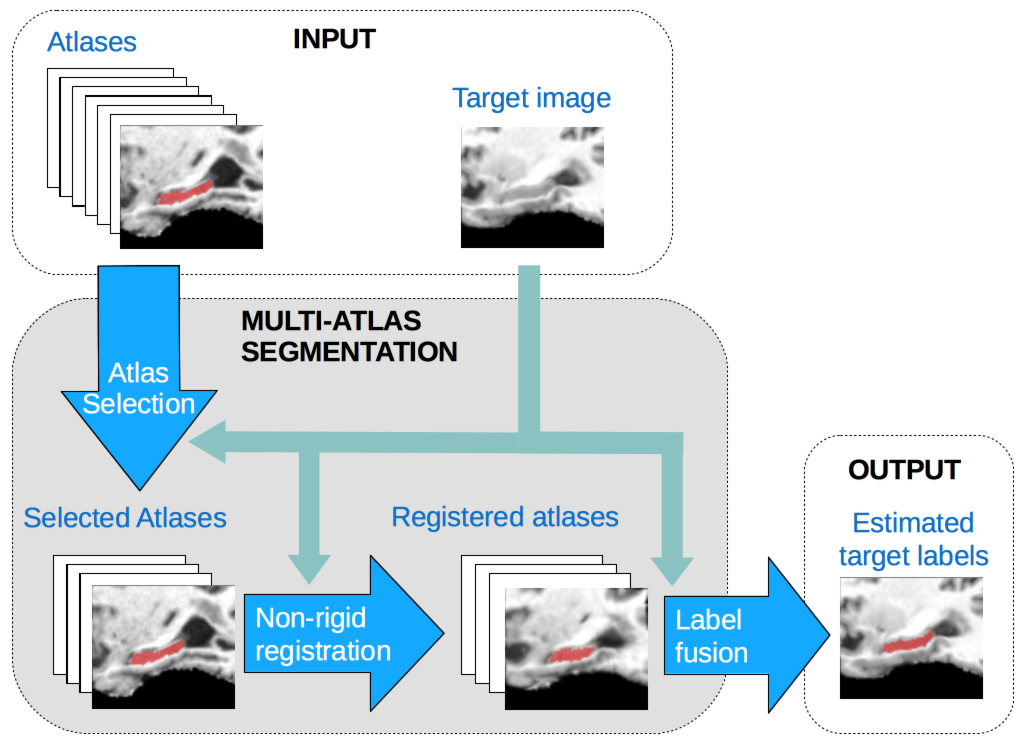
Our group is interested in the enhancement of MAS by the use of supervised learning techniques. Note that, supervised learning lends itself quite naturally to be used within MAS, since one of the pre-requisites of MAS is to have a training set of atlas images and labelmaps.
Our main past works consist in the development of a accurate atlas selection methods (Sanroma et al. TMI 2014), and the enhancement of patch-based label fusion (Sanroma et al. MedIA 2015, Wu et al. NeuroImage 2015).
In a recent work, we explore the use of discriminative dimensionality reduction techniques for patch-based label fusion. The idea is to transform the image patches to a lower-dimensional representation more suitable for similarity-based label fusion (Sanroma et al. ICML-MedIm 2015).
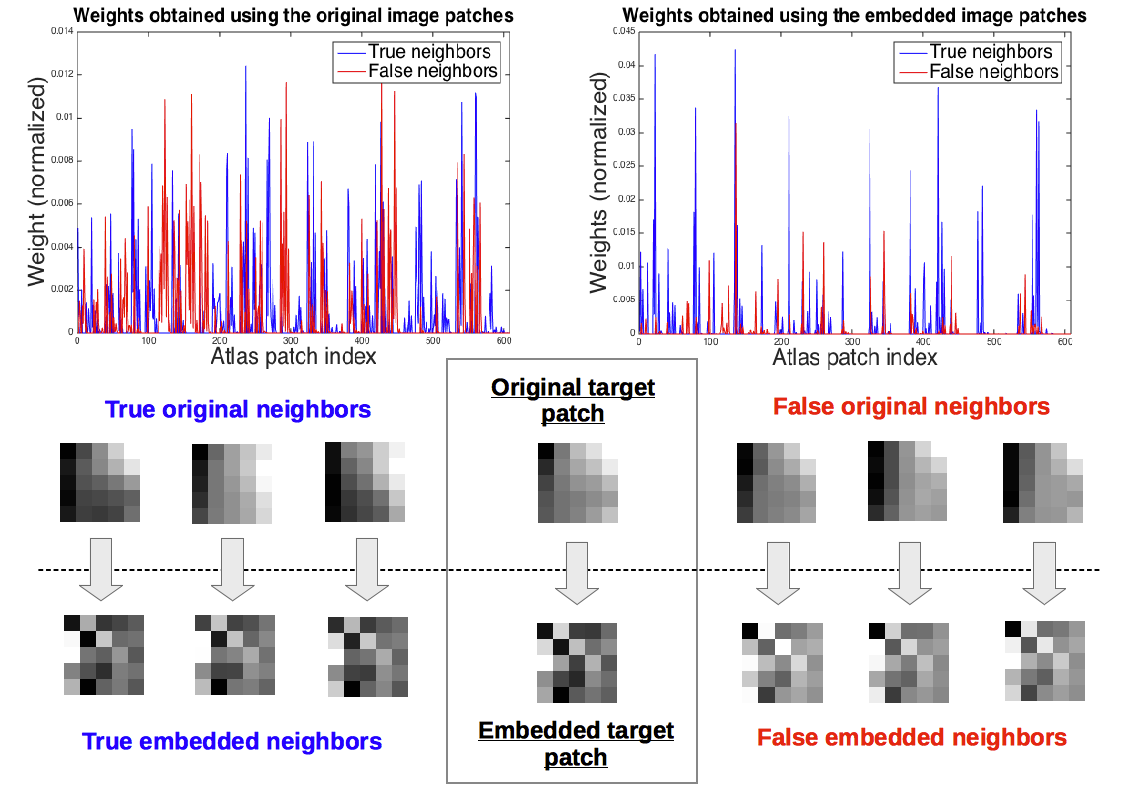
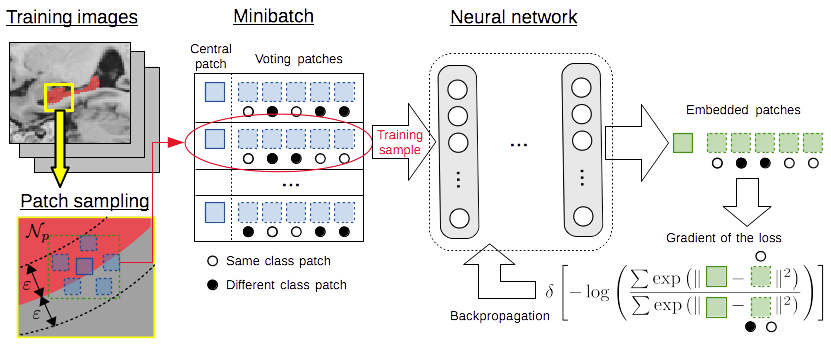
We have extended this approach to compute non-linear patch embeddings with deep neural networks. This paper is currently under review and will be linked here when published. Meanwhile, the code for this method can be accessed here.
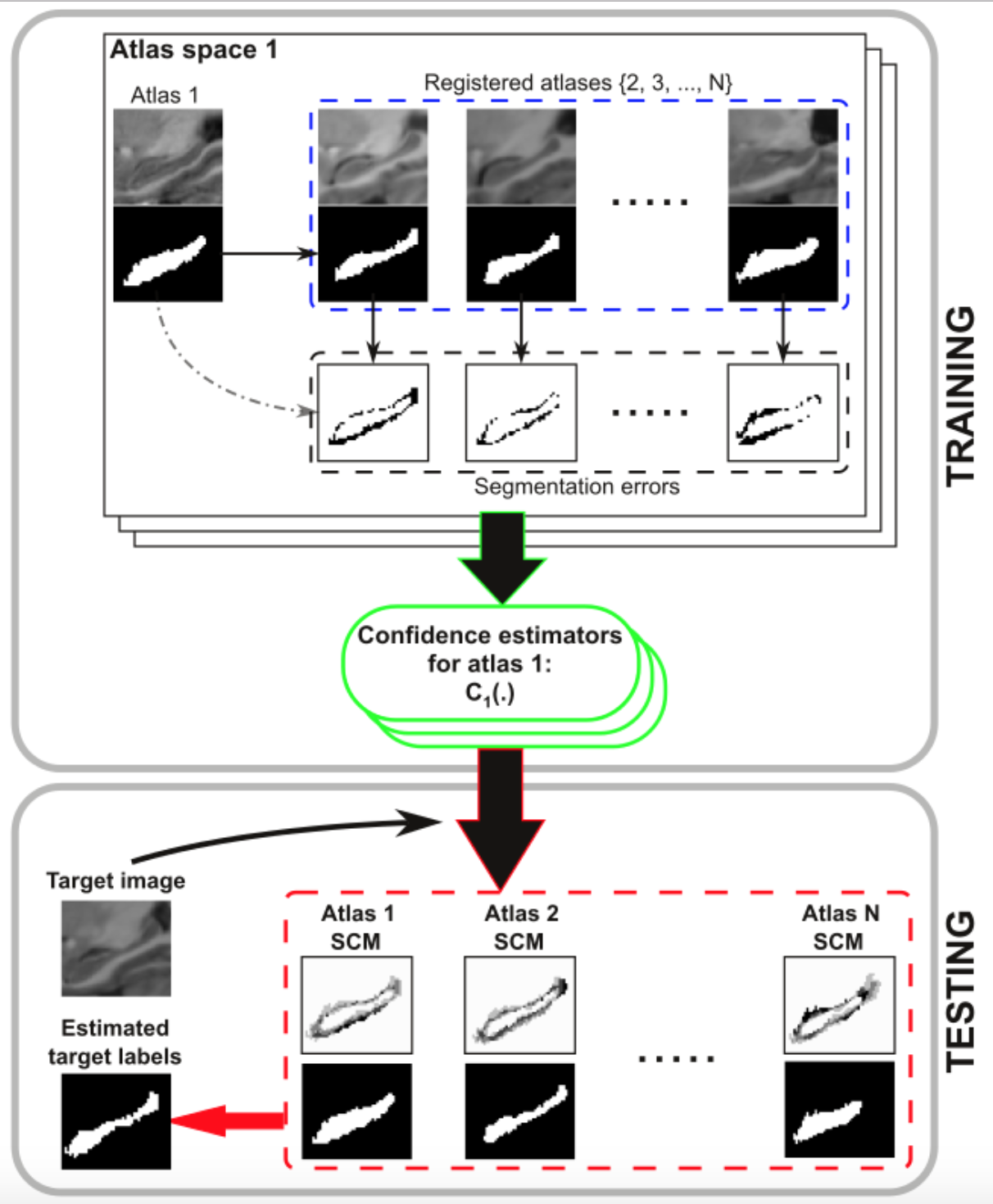
Estimation of the confidences for each atlas point is a key aspect in patch-based label fusion. We have proposed a method integrating discriminative classifiers into probabilistic multi-atlas segmentation for an optimal confidence estimation (O. M. Benkarim et al. MICCAI 2016).
Related to our work on early brain development, we have developed a method for segmenting structures and tissues of fetal and neonatal brain MRI (Sanroma et al. MLMI 2016). It features an ensemble classifier that learns the optimal combination of a set of pre-defined base methods. We obtained top-notch results in the NeoBrainS12 segmentation challenge.
Random walks with shape priors
xxx
Tree-based segmentation methods
xxx
Active Shape Models
xxx
