In network
7. In network
Proteomics: what proteins can tell us about history
Proteins are essential molecules for life. They have a variety of functions, from strengthening cells and tissues, to transporting substances to the organism, to defensive, storage and regulatory functions. When a protein does not function properly, it results in maladjustments that can lead to disease.

Proteomics is the large-scale study of proteins and their role in organisms. It offers insight into the functions of these molecules in health and disease. The UPF Proteomics Unit and the Centre for Genomic Regulation (CRG) work to determine exactly which proteins there are in a sample, in what amounts, what their function is, and when and where they carry it out. According to Eduard Sabidó, head of the unit, ‘To answer these questions, we use mass spectrometry. This technique allows us to measure thousands of proteins from a cell, which gives us a global view. Mass spectrometry is like a molecular scale that we use to weigh the proteins. Based on the weight, we can deduce the type of protein. If we were weighing the members of a family and we saw that one of them weighed 30 kg, we would be able to deduce that it was surely a child and not one of the parents. Likewise, we can use the proteins’ weight to determine what kind they are.’
Mass spectrometry is like a molecular scale that we use to weigh the proteins. Based on the weight, we can deduce the type of protein
The Proteomics Unit is made up of a team of ten people with different backgrounds: biologists, chemists, physicists and biochemists. The techniques they use require a multidisciplinary group, with people able to understand the biological mechanisms, in order to ask the right questions, and people with expertise in analytical chemistry and the physics of mass spectrometry, to properly interpret the results they obtain.

Mass spectrometry has many fields of application, from toxicology to water analysis or forensic studies. The members of the Proteomics Unit mainly focus on its biomedical applications. Within this field, they carry out basic research projects to answer basic questions about the action mechanisms of a cell or of organisms in general. They do these projects jointly with labs from various centres and institutions, both inside and outside the Barcelona Biomedical Research Park (PRBB), such as the Department of Experimental and Health Sciences (DCEXS) at UPF, the CRG or the Hospital del Mar Medical Research Institute (IMIM).
They also participate in more applied or translational biomedical projects, in collaboration with researchers from hospitals in Barcelona, including Hospital Vall d’Hebron, Hospital Germans Trias i Pujol, Hospital Clínic and Hospital Sant Joan de Déu. Hospitals have samples from patients who are very well characterized from a clinical perspective and have been medically monitored for long periods of time. For example, they have information about the early symptoms and diagnosis of a cancer, the treatment followed, and whether or not it was effective years later. ‘We re-analyse those samples, the biopsies obtained at the first visit, and ask whether there are any proteins that can help us determine which treatment would work best for that person. It does not have an immediate healthcare application, but it is clinical research that can provide us with important information to improve the healthcare system of tomorrow’, Sabidó explained.
He continued, ‘We can do these types of studies now because proteomics has been solidifying as an analytical technique in biomedicine for many years, with applications that have become routine and very robust. Because of this development, in recent years we have been able to apply the technique to highly promising new fields.’ In this line, the team have recently collaborated on several projects with experts from very different areas of the university, including terminology and linguistics, archaeology and evolutionary biology.
Proteomics has been solidifying as an analytical technique in biomedicine for many years, with applications that have become routine and very robust
Palaeoproteomics, the study of samples so old they cannot be analysed through DNA
Researchers from the Institute of Evolutionary Biology (IBE: CSIC-UPF) have been working with the Proteomics Unit to use proteomics to study evolution in hominids. Thanks to the resistance and high degree of conservation of some proteins, their analysis can be used to study very old samples that cannot be analysed using DNA sequencing techniques. This is because, depending on the type of sample and site, the DNA, which is very sensitive to environmental conditions, can be completely degraded.
Thanks to the resistance and high degree of conservation of some proteins, their analysis can be used to study very old samples that cannot be analysed using DNA sequencing techniques
The IBE and Proteomics Unit researchers recently participated in a study published in Nature led by the University of Copenhagen (Denmark), in collaboration with the National Research Centre for Human Evolution (CENIEH-ICTS) in Burgos (Spain). In that project, mass spectrometry was used to sequence ancient proteins in the enamel of an 800-thousand-year-old Homo antecessor tooth to confidently determine the position of the species in the human family tree. Their findings provided evidence for the idea that Homo antecessor was a sister group to the group of hominids containing Homo sapiens, Homo neanderthalensis and Homo denisovans.
According to Tomàs Marquès-Bonet, director of the IBE and co-author of the paper, ‘The recovery of molecular samples from ancient fossils combined with new computational techniques has revolutionized our understanding of the relationships between species that are no longer with us. The next few years will be fascinating. We are just now starting a five-year European Training Networks (ETN) project, called Palaeoproteomics to Unleash Studies on Human History (PUSHH), with many of the paper’s co-authors from different leading European institutions. The aim is to determine at the molecular level the ancestors of the Homo genus and beyond.’
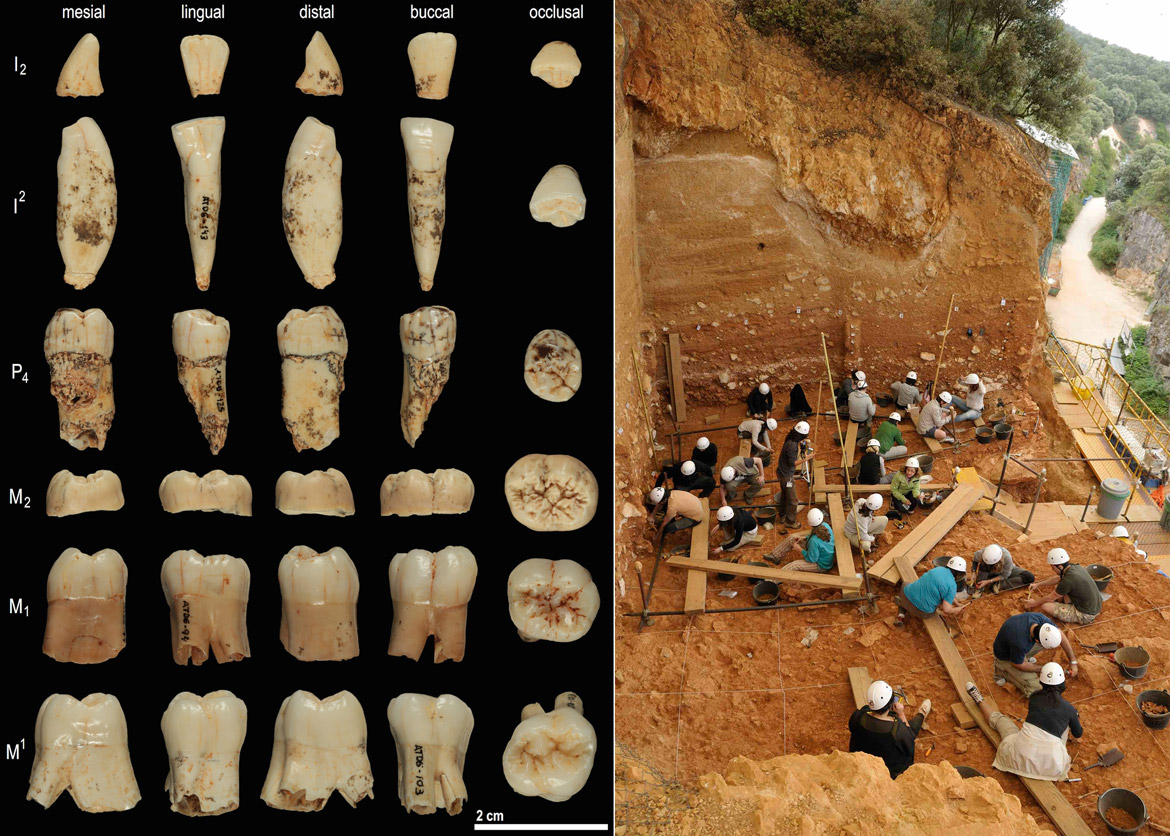
‘In 1997, researchers at Atapuerca opened an enormous scientific debate about the role of Homo antecessor; we have now closed it from a place that no one could have imagined then, palaeoproteomics. We hope that the same technique can be used to close many more debates in the future’, added Carles Lalueza-Fox, principal investigator at the IBE and a co-author of the paper.
‘Our participation in this project was very encouraging. It is a very new field of application of proteomics. Personally, I find it fascinating that we can analyse samples that are a million years old and extract relevant information to better explain the origin of our species. We participated in this project in collaboration with researchers from Denmark and Germany, but in the future, we would like to be able to carry out these kinds of projects independently from Barcelona’, explained Cristina Chiva, the Proteomics Unit scientist who was most involved in the project.
The application of proteomics to explore archaeological questions
Based on the palaeoproteomics project, the Proteomics Unit has delved into an adjacent field, archaeology, through collaboration with researchers from the Culture and Socio-Ecological Dynamics (CaSEs) research group from the Department of Humanities at UPF. ‘We are preparing this project and assessing its potential. It is a good example of a cross-disciplinary application that goes beyond the field of biomedicine’, said Sabidó.
According to Marco Madella, an ICREA-UPF research professor and coordinator of the CaSEs group, ‘The use of proteomics is increasingly important in archaeology. A number of groups around the world have developed the application of this technique to explore archaeological questions.’
The collaboration will take place within the Modelling the Agricultural Origins of South Asia - ModAgrO project, which aims to understand the dynamics of the origin of agriculture in the Indus River Valley and the subsequent development of one of the world’s earliest prehistoric urban civilizations, the Indus Civilization (2600-1900 BCE), in South Asia, in modern-day India and Pakistan. It is a joint project between Shah Abdul Latif University and UPF, with funding from the Palarq Foundation.
According to Madella, ‘First, along with bone anatomy, proteomics can help us determine the types of animals domesticated by the Indus Valley civilization and, more specifically, understand the frequency and proportion of sheep to goats.’ This is important because the two animals occupy very different ecological niches and, thus, can shed light on this ancient civilization’s land use strategies and environmental impact.
The use of proteomics to analyse protein remains on potsherds can aid in the study of the Indus civilization diet
Second, the use of proteomics to analyse protein remains on potsherds can aid in the study of the Indus civilization diet, how they processed agricultural products to make food. By analysing the organic remains of tools found at archaeological sites, such as different vessels, it is possible to determine what type of food they had, for example, whether they ate wheat or drank milk.

The terminology of proteomics
The collaboration between the IULATERM research group from the UPF Department of Translation and Language Sciences and the Proteomics Unit began through the optional subject Terminology Management, shared by the Translation and Interpreting and Applied Languages bachelor’s degrees. The seminars are organized around projects to develop and transfer multilingual vocabularies on specialized topics. Each seminar develops a previously unpublished vocabulary of between 100 and 200 terms, from the search for specialized documents, the compilation of corpora and the extraction of terms to the preparation of terminological files and, finally, the publication of a digital edition of the vocabulary.
According to Mercè Lorente, principal investigator of the IULATERM group and director of the Institute for Applied Linguistics (IULA), ‘In the 2018-2019 academic year, one of the vocabularies we set out to do was a bioinformatics vocabulary, since the official terminology centre of Catalonia, TERMCAT, had suggested that the subject had yet to be addressed in depth from a terminological point of view.’ However, the group soon learnt that bioinformatics is used to study massive data sets on several aspects of biomedicine, including genomics, transcriptomics, proteomics, etc. ‘When we saw how broad the discipline was, we decided to focus on just one of the associated branches’, she explained.

‘Following a search, we decided that proteomics was a rapidly evolving field that lacked sufficient terminological resources for both students and professionals’, said Laia Vidal Sabanés, the professor in charge of this vocabulary. ‘The Proteomics Unit confirmed our observation and agreed to collaborate. In the first phase of work, they helped us develop the field tree [a conceptual schema of the vocabulary] and with the basic bibliographical search’, she continued.
Proteomics was a rapidly evolving field that lacked sufficient terminological resources for both students and professionals
The students’ initial work is followed by a methodological, linguistic and lexicographical quality review. This academic year, they finished reviewing the methodological and linguistic quality of the vocabulary, and researchers from the Proteomics Unit reviewed the nomenclature (to identify any missing terms) and definitions (to ensure they properly reflect the concepts). ‘We are currently entering the last changes before completing the vocabulary with the final publication stage. We hope to publish the electronic dictionary when the lockdown is over’, said Mariona Arnau, the IULA specialist who did the review.
‘Participating in a language project was a challenge and required us to change how we think. In some cases, the concepts we had to define did not even have names in Catalan, so we had to create them’, said Chiva.

Within this framework of interaction between teaching and development and transfer projects, four multilingual vocabularies related to biomedicine have been published so far: Vocabulari bàsic del genoma humà [Basic vocabulary of the human genome] (2007), in collaboration with the REALITER network; Vocabulari de les malalties minoritàries [Vocabulary of minority diseases], linked to the JUNTS project; Vocabulari multilingüe de càncer de mama per a pacients [Multilingual breast cancer vocabulary for patients], in collaboration with the Catalan Oncology Institute (ICO) (forthcoming); and, finally, another vocabulary related to the health sciences, Vocabulari de nutrició pública [Vocabulary of public nutrition], in collaboration with the Office Québecois à la Langue Française (OQLF).
The future challenges of proteomics
Several fields of great biomedical interest pose major analytical challenges. According to Sabidó, ‘Being able to perform routine screening of large populations, as is already done in some cases in genomics, is one major challenge. You need to be able to process a lot of samples in a short time, to analyse cohorts of thousands or tens of thousands of patient samples.’
Another challenge is to narrow the gap between more clinical proteomics and actual clinical diagnosis. Most detection kits today are made for antibodies, and there are many limitations in terms of sensitivity or specificity. ‘Even though these limitations could be overcome with mass spectrometry, this technique for analysing proteins is not yet used in hospitals. Many people in the field are working to advance in this direction and improve tests and diagnosis’, he explained.
In basic biomedical research, analysing individual cells, as is also done in genomics, is another challenge. That would make it possible to see the variability between the proteomes of different cells, for example, of a tumour, where not all cells are exactly the same nor do they all behave the same way.

The importance of interdisciplinary collaboration
According to Vidal Sabanés, ‘We really value this multidisciplinary experience. We have been using this collaborative terminology system for years and consider interaction with expert groups to be very enriching for terminology projects. They help us immerse ourselves in the subject, allow us to get to know their language resource needs first hand and assure the quality of our output. Most importantly, these interactions are a magnificent personal exchange experience.’

‘At universities in general, including ours, we often talk about the benefits of interdisciplinarity, but, unfortunately, we do not always practice what we preach. We are certainly not the only ones, but we believe we are pioneers in these practices and we continue to be staunch advocates of them’, added Lorente.
The collaboration between the different disciplines affords each field added value that we could not otherwise achieve
With regard to archaeology, Madella stressed, ‘It is by nature an inter- and transdisciplinary field. Being able to cover multiple areas of knowledge is essential to our research.’ Sabidó concluded, ‘The collaboration between the different disciplines is very interesting, because we complement each other. It affords each field added value that we could not otherwise achieve.’
